Saving CP9/19
At Absolute Antibody, we specialise in the sequencing, engineering, and recombinant expression of antibodies and Fc Fusion proteins. We pride ourselves in going the extra mile to help our customers realise their projects, often taking on difficult challenges and apparent ‘lost causes’.
This is one such story of some work we did with Ximbio – the research tools commercialisation business of Cancer Research UK.
| Antibody Product Code | Species / Format |
|---|---|
| Ab00612-7.1 | Rat IgG2a (original format) |
| Ab00612-7.4 | Rat IgG2a (Fc Silent) |
| Ab00612-23.0 | Rabbit IgG |
In November 2015 we were approached by Ximbio, who were concerned about their popular anti-cisplatin-modified-DNA antibody CP9/19, invented by Mike Tilby at Newcastle University. They believed there may have been historical contamination in their hybridoma, and attempts to source back to a clonal stock had failed.
The original hybridoma was a rat IgG2a, but we detected only mouse antibody in the culture using a standard isotyping kit. We used a PCR-based sequencing approach at the time, and so designed a strategy to suppress the mouse sequences and amplify any remnants of the rat heavy and light chain sequences.
All attempts were unsuccessful, with over 50 clones sequenced. At this point, many organisations would be ready to abandon the project. But Ximbio had detected some limited CP9/19 activity from the hybridoma, meaning the rat cells were in there somewhere. So we didn’t give up.
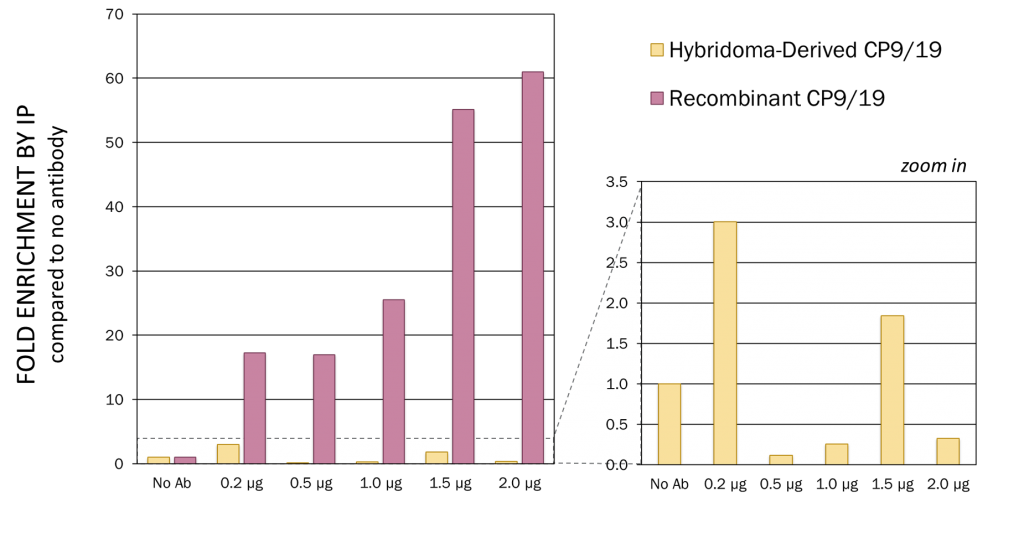
Figure 1. Recombinant CP9/19 showed significantly improved performance compared to hybridoma-derived CP9/19 from a leading commercial competitor. Antibody titration: Immunoprecipitation IP. Based on qPCR on MFA2 locus in yeast genome. LEFT: Recombinant CP9/19 showing a much higher fold enrichment in comparison to hybridoma-derived CP9/19, including evidence of dose-dependent performance in the recombinant version. RIGHT: Scaled to show poor performance of hybridoma-derived CP9/19. This is attributed to contamination of the original hybridoma culture.
We wondered if we could somehow enrich for the rat hybridoma. As fusions of myeloma and lymphoid cells, hybridomas should express the leukocyte common antigen CD45. Perhaps we could use a mouse-anti-rat CD45 antibody to select for the elusive rat cells expressing CP9/19? The thinking paid off, and we obtained a rat sequence for the antibody heavy chain.
The light chain, however, still appeared to be from a mouse. This is where our in-house expertise saved the day again. After some scrutiny we identified the “mouse” sequence as an unusual rat allele. Without a high-level of familiarity with antibody sequences and structure, this could easily have gone unnoticed.
With a sequence to work with, we could express CP9/19 using our recombinant platform, but we could not test it ourselves as cisplatin is a carcinogen we were not equipped to work with. The data shown here were obtained by Simon Reed and his team at Cardiff University.
The story of this project illustrates the expertise and tenacity that we apply to all of our customers’ projects at Absolute Antibody.
Chris Birchall, who was part of Ximbio’s portfolio team at the time of this project, said:
“The antibody CP9/19 was first characterised almost 30 years ago. CP9/19 was widely adopted by the DNA damage research community and has been applied in more than 15 publications. It was of grave concern to Ximbio when we learnt that the rat hybridoma cells were contaminated with mouse antibody producing cells which were much more proliferative but as yet unknown. The Absolute Antibody team excelled in this project, firstly to ensure the longevity of this antibody through sequencing and secondly to enable the generation of an antibody preparation that is superior in performance to one derived from the hybridoma”
Contact us with your own challenge – we’d love to hear from you.
Read more about our sequencing services here.
Latest News
Upcoming Events
Please join us at the following conferences and events. Stop by our booth, or get in touch to arrange a meeting.
See All Dates
 United Kingdom (UK)
United Kingdom (UK)