In simplistic terms antibodies perform two main functions in different regions of their structure. While one part of the antibody, the antigen binding fragment (Fab), recognizes the antigen, the other part of the antibody, known as the crystallizable fragment (Fc), interacts with other elements of the immune system, such as phagocytes or components of the complement pathway, to promote removal of the antigen.
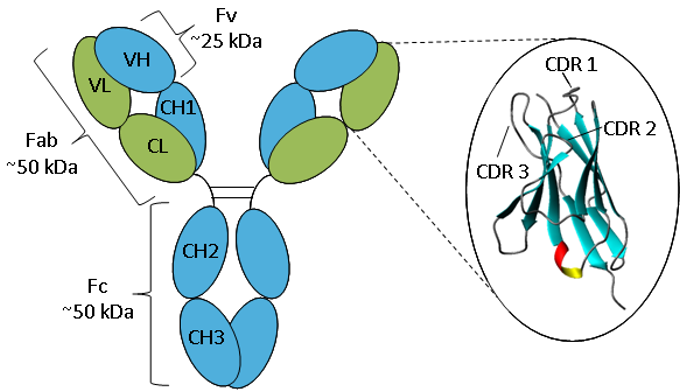
Figure. Schematic representation of an IgG.
An antibody consists of two heavy chains (blue) and two light chains (green) folded into constant and variable domains. The enlargement of the variable domain shows a ribbon representation of the β-sheet framework and CDR loops.
Antibodies all have the same basic structure consisting of two heavy and two light chains forming two Fab arms containing identical domains at either end attached by a flexible hinge region to the stem of the antibody, the Fc domain, giving the classical ‘Y’ shape. The chains fold into repeated immunoglobulin folds consisting of anti-parallel β-sheets (1), which form either constant or variable domains. The Fab domains consist of two variable and two constant domains, with the two variable domains making up the variable fragment (Fv), which provides the antigen specificity of the antibody (2) with the constant domains acting as a structural framework. Each variable domain contains three hypervariable loops, known as complementarity determining regions (CDRs), evenly distributed between four less variable framework (FR) regions. It is the CDRs that provide a specific antigen recognition site on the surface of the antibody and the hypervariability of these regions enables antibodies to recognize an almost unlimited number of antigens (3).
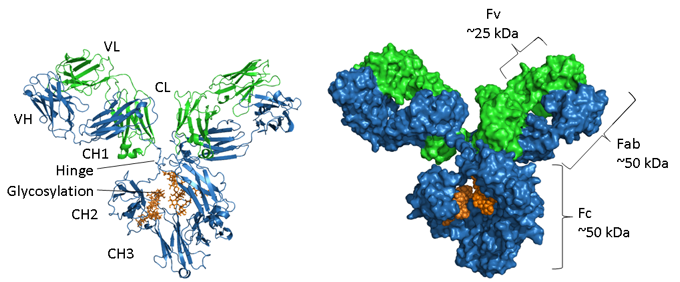
Figure. Structural representations of an IgG.
The heavy chain is shown in blue, light chain in green and glycosylation in orange. On the left is a ribbon representation showing the secondary structure elements and on the right hand side is a space-filled model of the same molecule. PDB accession number of the mouse IgG1 is 1IGY.
Antibodies are glycosylated proteins, with the position and extent of glycosylation varying between isotypes. As displayed in the image above the Fc region of an IgG consists of two paired CH3 domains and, in contrast, two CH2 domains that are separated and do not interact but have two oligosaccharide chains interposed between them. These chains cover the hydrophobic faces that would normally lead to domain pairing. The N-glycans contain a common core region of two N-acetyl-glucosamine residues (GlcNAc) linked to an asparagine (N297 in human IgG1) via an amide bond and three mannose residues. This core structure may contain additional terminal sugars, such as mannose, GlcNac, galactose, fucose and sialic acid, generating a large amount of heterogeneity (4).
<< A brief history of antibodies Antibody isotypes & subtypes >>
References
- Poljak, R.J., Amzel, L.M., Avey, H.P., Chen, B.L., Phizackerley, R.P., and Saul, F. (1973). Three-dimensional structure of the Fab’ fragment of a human immunoglobulin at 2,8-A resolution. Proc. Natl. Acad. Sci. U.S.A. 70, 3305–3310.
- Inbar, D., Hochman, J., and Givol, D. (1972). Localization of antibody-combining sites within the variable portions of heavy and light chains. Proc. Natl. Acad. Sci. U.S.A. 69, 2659–2662.
- Morea, V., Lesk, A.M., and Tramontano, A. (2000). Antibody modeling: implications for engineering and design. Methods 20, 267–279.
- Wright, A., and Morrison, S.L. (1998). Effect of C2-associated carbohydrate structure on Ig effector function: studies with chimeric mouse-human IgG1 antibodies in glycosylation mutants of Chinese hamster ovary cells. J. Immunol. 160, 3393–3402.

 United Kingdom (UK)
United Kingdom (UK)